|

PhD Student

Project Leader
|
|
Intrahepatic cholestasis describes defective bile formation within hepatocytes and comprises a heterogeneous group of liver diseases ranging from rare inherited chronic forms of disease to more common acquired and sometimes even transient forms. The underlying molecular mechanisms comprise defects in bile acid synthesis, nuclear signaling, vesicular trafficking, canalicular transport of bile acids and bile constituents across the epithelial barrier, tight junction-mediated impaired barrier function, and maintenance of cell polarity resulting in impaired bile secretion from hepatocytes into the canalicular lumen.
The main purpose of this project is the identification and characterization of genetic variants underlying hereditary forms of intrahepatic cholestasis, which are characterized by defective barrier function either in the hepatocyte or cholangiocyte using next generation sequencing (NGS) approaches. Therefore, the focus lies on the six best-known cholestasis-associated genes ATP8B1, ABCB11, ABCB4, TJP2, NR1H4, MYO5B, and other recently identified genes as SEMA7A, KIF12, ABCC12, USP53, ZFYVE19, FGFR4. In patients without relevant variants in these genes, other possibly related genes may be identified to be responsible for genetically determined cholestasis. Moreover, this project represents the basis for choosing variants or patients for further analysis in silico and in vitro analysis.
Another part of the project is to determine whether poly- or oligogenic variant combinations may induce cholestatic phenotypes. Genetic variants associated with other chronic hepatopathies (in HFE, SERPINA1 or genes related to metabolic liver disease) may contribute to manifestation and progression of liver disease in patients with hereditary cholestasis. Thus, another aim is to compile genetic information of the known relevant cholestasis-related genes including other specific variants for each patient to define a mutational burden using HiChol registry data.
The achieved results will lead to an improved patient cohort characterization, to the detection of novel variants in the known as well as novel cholestasis-related genes and a better understanding of the contribution of different genes to the overall phenotype. The obtained knowledge will improve health care for this rare disease with respect to diagnosis through establishment of quick and reliable genetic testing, disease monitoring (HCC risk of specific mutation subclasses) and more directed therapeutic approaches.
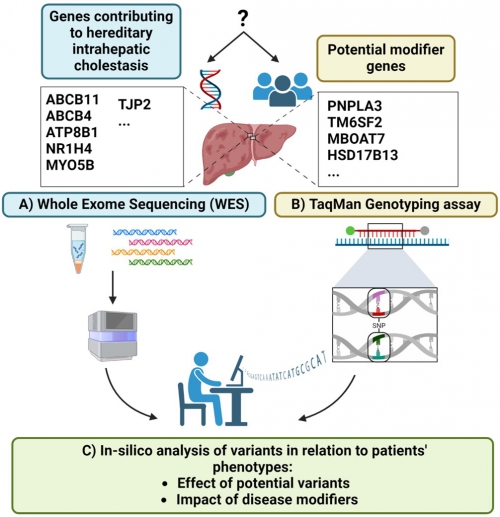
Workflow for genetic analysis in hereditary intrahepatic cholestasis. Variants in genes involved in bile formation are related to hereditary intrahepatic cholestasis. Modifier variants may contribute to disease severity and progression. A) Whole exome sequencing (WES) is performed to detect potentially relevant genetic variants in the protein-coding and adjacent regions. B) TaqMan genotyping assay is used to identify risk-modifying single nucleotide polymorphisms (SNPs) that may influence disease manifestation. C) Bioinformatic analysis of WES results is carried out for alignment, annotation, and variant prioritization.
|