Shivani Singh

Shivani Singh
PhD Student
Project 11-3
P11-3: Optimized models for studying intra- and intercellular crosstalk in biliary diseases
Shivani SinghPhD Student
Verena Keitel-AnselminoProject Leader |
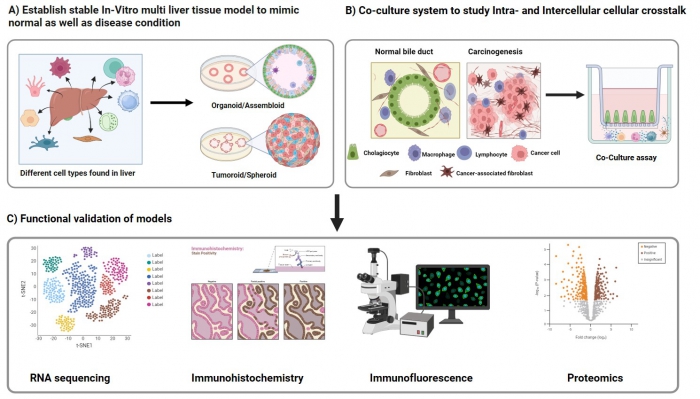
Biliary diseases are characterized by complex interactions between various liver cell types, including hepatocytes, cholangiocytes, and immune cells, mediated through intricate intra- and intercellular signalling pathways. Understanding these communication networks is essential for deciphering disease mechanisms and identifying effective therapeutic targets. Traditional models often fail to capture the complexity of cellular crosstalk and the dynamic microenvironment of the biliary system, limiting their utility for understanding disease progression and testing therapies. This highlights the need for more sophisticated multicellular organoid models and patient-specific systems to enable personalized medicine. This study focuses on the development and optimization of advanced in vitro models to investigate cellular crosstalk in biliary diseases. Key approaches include:
The advanced models developed in this study will enhance our understanding of biliary disease pathophysiology and provide valuable platforms for drug screening and the development of targeted therapies. This work underscores the importance of precise and physiologically relevant models in advancing biliary disease research and lays a solid foundation for future translational studies.
Establishment of in vitro liver models using organoids, spheroids, and co-culture systems to study cellular crosstalk in normal and diseased conditions. Functional validation is performed through RNA sequencing, immunohistochemistry, immunofluorescence, and proteomics. |
Photos: by UMMD, Melitta Schubert/Sarah Kossmann